|
|
|
|
|
黑曲霉的代谢网络重建 | BMC Journals |
|
|
论文标题:A community-driven reconstruction of the Aspergillus niger metabolic network
期刊: Fungal Biology and Biotechnology
作者:Julian Brandl, Maria Victoria Aguilar-Pontes, Paul Schäpe, Anders Noerregaard, Mikko Arvas, Arthur F. J. Ram, Vera Meyer, Adrian Tsang, Ronald P. de Vries and Mikael R. Andersen
发表时间:2018/09/26
数字识别码:10.1186/s40694-018-0060-7
原文链接:https://fungalbiolbiotech.biomedcentral.com/articles/10.1186/s40694-018-0060-7?utm_source=WeChat&utm_medium=Social_media_organic&utm_content=DaiDen-BMC-Fungal_Biology_and_Biotechnology-Biotechnology-China&utm_campaign=AJL_USG_BSCN_DD_AspergillusNiger_Wechat
微信链接:https://mp.weixin.qq.com/s/EERAHTMvj2ucdqnWc_DVEQ
原文作者:Julian Brandl, Maria Victoria Aguilar-Pontes, Paul Schäpe, Anders Noerregaard, Mikko Arvas, Arthur F. J. Ram, Vera Meyer, Adrian Tsang, Ronald P. de Vries and Mikael R. Andersen
很多生物的实验数据都分散在数百篇出版物和多个数据储存库中。要想对生物代谢有系统的认识,就必须将数据整合到统一的知识网络中。黑曲霉(Aspergillus niger)是酶和酸生产工业中使用的重要真菌。为了对黑曲霉进行合理的代谢改造,可以收集现有的资料并在基因组水平模型中加以整合,进而设计出能够改善黑曲霉宿主性能的有效方案。

近期发表于Fungal Biology and Biotechnology的一篇文章“A community-driven reconstruction of the Aspergillus niger metabolic network”描述了来自丹麦技术大学的Mikael R. Andersen及其研究团队根据876篇文献中的信息更新了现有的黑曲霉代谢模型,将模型覆盖范围扩展到了940种反应、777种代谢物和454个基因。他们把来自论文和专利的实验数据以及自己的实验整合成一致的网络,最终确定了黑曲霉iJB1325基因组的水平模型。信息的纳入采用了标准化方式,以便将来能进行自动化测试和持续改进。这一实验数据库共有471个测试案例,而iJB1325模型符合其中的373个。研究者们还重新分析了现有的转录组学和生理学定量数据以进一步探索黑曲霉的代谢。此外,该模型包含了对模型结构的3,482次检查,是迄今为止最有力的黑曲霉经基因组水平模型。研究者们进一步利用现有数据构建了ATCC 1015和CBS 513.88的菌株特异性模型,用户可根据实验数据对最新模型进行验证。更新后的模型符合SBML标准,因此用户可通过自己的惯用软件轻松模拟该模型。
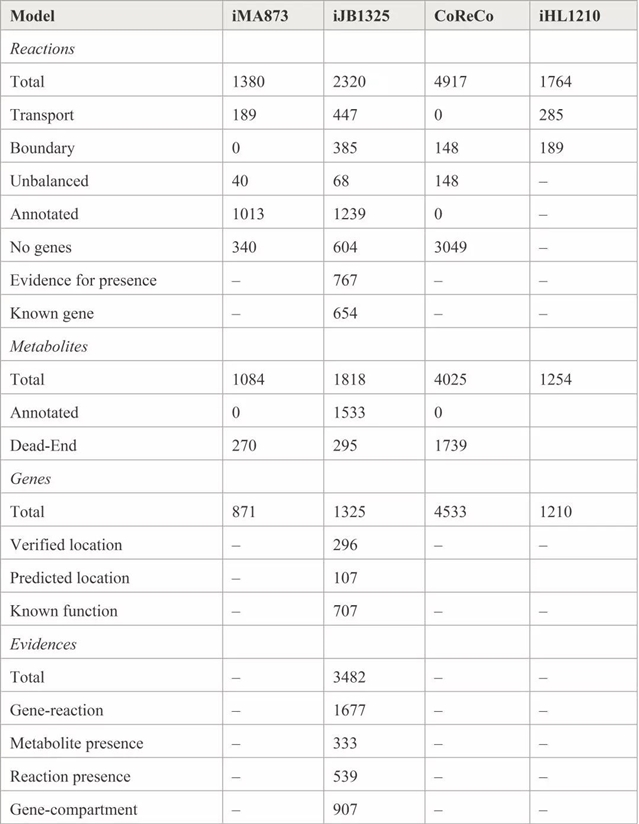
图:表格描述了不同模型的主要数据
本文介绍的黑曲霉iJB1325模型将可用数据整合到高度规整的基因组水平模型中,一方面便于模拟物质/能量流动分布,另一方面也为基因组水平数据的解读提供了代谢学背景。
点击阅读全文了解详情:
摘要:
Background
Aspergillus niger is an important fungus used in industrial applications for enzyme and acid production. To enable rational metabolic engineering of the species, available information can be collected and integrated in a genome-scale model to devise strategies for improving its performance as a host organism.
Results
In this paper, we update an existing model of A. niger metabolism to include the information collected from 876 publications, thereby expanding the coverage of the model by 940 reactions, 777 metabolites and 454 genes. In the presented consensus genome-scale model of A. niger iJB1325 , we integrated experimental data from publications and patents, as well as our own experiments, into a consistent network. This information has been included in a standardized way, allowing for automated testing and continuous improvements in the future. This repository of experimental data allowed the definition of 471 individual test cases, of which the model complies with 373 of them. We further re-analyzed existing transcriptomics and quantitative physiology data to gain new insights on metabolism. Additionally, the model contains 3482 checks on the model structure, thereby representing the best validated genome-scale model onA. niger developed until now. Strain-specific model versions for strains ATCC 1015 and CBS 513.88 have been created containing all data used for model building, thereby allowing users to adopt the models and check the updated version against the experimental data. The resulting model is compliant with the SBML standard and therefore enables users to easily simulate it using their preferred software solution.
Conclusion
Experimental data on most organisms are scattered across hundreds of publications and several repositories.To allow for a systems level understanding of metabolism, the data must be integrated in a consistent knowledge network. The A. niger iJB1325 model presented here integrates the available data into a highly curated genome-scale model to facilitate the simulation of flux distributions, as well as the interpretation of other genome-scale data by providing the metabolic context.
阅读论文全文请访问:
https://fungalbiolbiotech.biomedcentral.com/articles/10.1186/s40694-018-0060-7?utm_source=WeChat&utm_medium=Social_media_organic&utm_content=DaiDen-BMC-Fungal_Biology_and_Biotechnology-Biotechnology-China&utm_campaign=AJL_USG_BSCN_DD_AspergillusNiger_Wechat
期刊介绍:
Fungal Biology and Biotechnology(https://fungalbiolbiotech.biomedcentral.com/) is a peer-reviewed journal that publishes original scientific research and reviews covering all areas of fundamental and applied research which involve unicellular and multicellular fungi.
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。