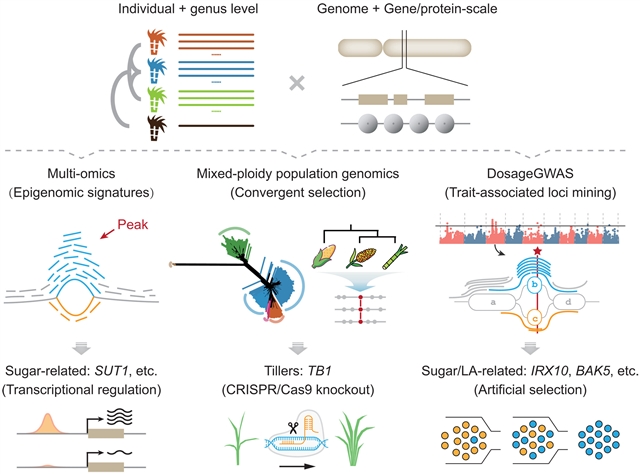
该课题组人员开发了一个基于多尺度图的泛基因组表示,将9个基因组组装成一个统一的参考,代表现代栽培品种和创始物种。每个同源(eo)的标志(包括同源和同源关系)染色体组保留47至57个单倍型和约74000至271000个基因等位基因。该框架支持多组学探索,包括同源(eo)日志系统和表观基因组特征。该泛基因组有助于对417个混合倍性甘蔗材料进行群体基因组学分析,揭示了趋同选择,并确定了与分蘖相关的Andropogoneae TB1同源物是提高甘蔗产量的有希望的基因编辑靶点。此外,泛基因组支持剂量信息全基因组关联研究,提高遗传力估计和鉴定糖或叶角相关位点,包括SaIRX10和SaBAK5。他们的分析框架为甘蔗和其他多倍体基因组的图谱遗传研究奠定了基础。
研究人员表示,甘蔗属特点具有复杂的基因组和不同的倍性水平。
附:英文原文
Title: Multiscale pangenome graphs empower the genomic dissection of mixed-ploidy sugarcane species
Author: Yumin Huang, Yixing Zhang, Qing Zhang, Gui Zhuang, Chunjia Li, Baiyu Wang, Ruiting Gao, Yi Xu, Yiying Qi, Xiuting Hua, Huihong Shi, Qiutao Xu, Wei Yao, Xinlong Liu, Yongwen Qi, Baoshan Chen, Muqing Zhang, Ray Ming, Haibao Tang, Jisen Zhang
Issue&Volume: 2026-02-05
Abstract: The sugarcane genus Saccharum is characterized by complex genomes with diverse ploidy levels. We developed a multiscale graph–based pangenome representation, which integrates nine genome assemblies into a unified reference, representing modern cultivars and founding species. Each homo(eo)logous (encompasses both homologous and homeologous relationships) chromosome set retains 47 to 57 haplotypes and ~74,000 to 271,000 gene alleles. This framework enables multiomics exploration, encompassing homo(eo)log systems and epigenomic signatures. The pangenome facilitates population genomics analyses of 417 mixed-ploidy Saccharum accessions, revealing convergent selection and identifying the Andropogoneae TB1 homolog linked to tillering as a promising gene-editing target to boost cane yield. Additionally, the pangenome supports dosage-informed genome-wide association study, improving heritability estimates and identification of sugar or leaf-angle–associated loci, including SaIRX10 and SaBAK5. Our analytical framework establishes a foundation for graph-based genetic studies in sugarcane and other polyploid genomes.
DOI: adx1616
Source: https://www.science.org/doi/10.1126/science.adx1616
