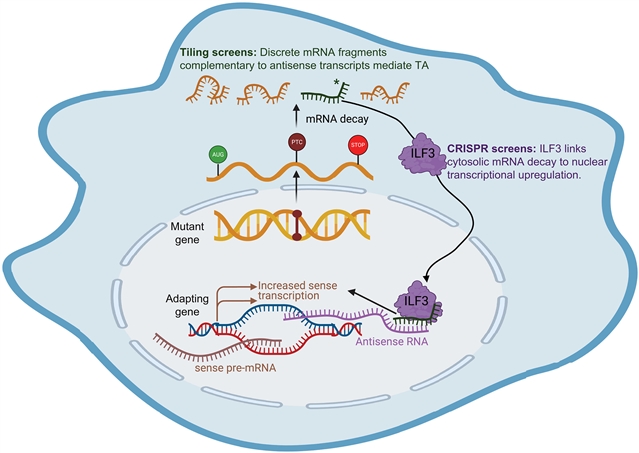
使用全基因组CRISPR筛选,课题组研究人员发现ILF3是一种RNA结合蛋白,在TA过程中连接细胞质mRNA衰变和转录,并表明它是一系列TA底物所必需的。ILF3富集于适应基因的RNA,通过dCas13人工募集ILF3可促进基因表达。使用平铺寡核苷酸筛选,该课题组人员确定触发RNA片段,激活适应基因时引入细胞。进一步的功能解剖揭示了触发序列和靶序列之间同源性的关键作用。这些发现增强了他们对TA的分子理解,并为基因表达增强的可编程寡核苷酸的设计提供了信息。
据介绍,转录适应(Transcriptional adaptation, TA)是一种遗传适应性机制,通过突变的信使RNA (mRNA)衰变诱导所谓的适应基因序列依赖的上调。胞质产生的mRNA片段如何影响核转录仍然知之甚少。
附:英文原文
Title: Mechanisms linking cytoplasmic decay of translation-defective mRNA to transcriptional adaptation
Author: Mohamed A. El-Brolosy, Atharv Oak, An T. Hoang, Yassine Damergi, André Fischer, Reuben A. Saunders, Jingchuan Luo, Amer Balabaki, Jeremy Guez, Troy W. Whitfield, Seth R. Goldman, Arash Latifkar, Yuancheng Ryan Lu, Didier Y. R. Stainier, Konrad J. Karczewski, Olivia Corradin, Jonathan S. Weissman
Issue&Volume: 2026-02-12
Abstract: Transcriptional adaptation (TA) is a genetic robustness mechanism through which mutant messenger RNA (mRNA) decay induces sequence-dependent up-regulation of so-called adapting genes. How cytoplasmically generated mRNA fragments affect nuclear transcription remains poorly understood. Using genome-wide CRISPR screens, we uncover ILF3 as an RNA binding protein connecting cytoplasmic mRNA decay and transcription during TA and show that it is required for a range of TA substrates. ILF3 is enriched at adapting genes’ RNAs, and its artificial recruitment through dCas13 promotes gene expression. Using tiling oligonucleotide screens, we identify trigger RNA fragments that activate adapting genes when introduced into cells. Further functional dissection reveals a critical role for homology between trigger and target sequences. These findings enhance our molecular understanding of TA and inform the design of programmable oligonucleotides for gene expression augmentation.
DOI: aea1272
Source: https://www.science.org/doi/10.1126/science.aea1272
