近日,中国科学院深圳先进技术研究院金帆团队报道了解码频率调制信号增加了细菌第二信使网络的信息熵。相关论文于2025年9月15日发表在《自然—物理学》杂志上。
细菌第二信使网络通过振幅和频率调制策略传输环境信息。然而,细胞解码频率编码信号的机制仍然知之甚少。
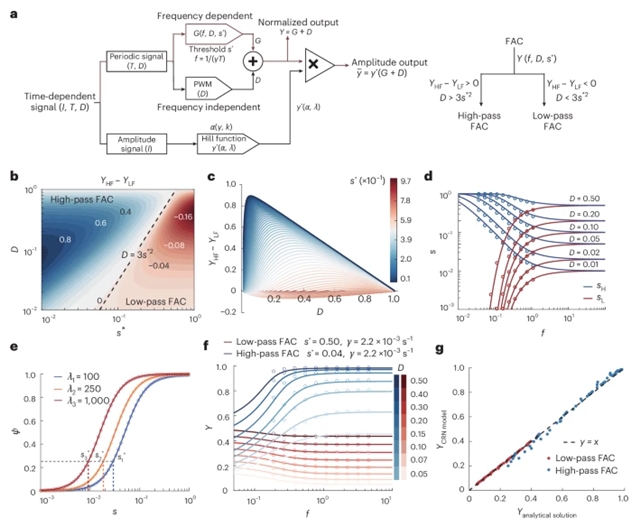
通过重建铜绿假单胞菌的环磷酸腺苷第二信使系统,研究组证明了频率到幅度的信号转换通过三个不同的滤波模块出现,这些模块将频率编码的信号解码为基因表达模式。他们的数学框架预测了由无量纲阈值参数控制的频率滤波范围。研究组验证了这些主题合成电路和自动化实验平台。定量分析表明,在给定的参数条件下,频率调制比单独的幅度调制更显著地扩展了可访问的状态空间。与仅振幅控制的0.8次幂相比,可访问状态的总数与频率增强控制的受调节基因数量的平方成正比。
当使用基于频率的控制时,这会在三个基因系统中产生大约两个额外的信息熵比特。该发现确立了细菌第二信使网络中基于频率的信号处理的基本原理,揭示了细胞如何利用时间动力学来调节多个基因并扩展可访问的状态空间。这为细胞信息物理学和合成生物学的设计原理提供了见解。
附:英文原文
Title: Decoding frequency-modulated signals increases information entropy in bacterial second messenger networks
Author: Zhang, Rongrong, Wan, Shengjie, Xiong, Jiarui, Ni, Lei, Li, Ye, Huang, Yajia, Li, Bing, Li, Mei, Yang, Shuai, Jin, Fan
Issue&Volume: 2025-09-15
Abstract: Bacterial second messenger networks transmit environmental information through both amplitude and frequency modulation strategies. However, the mechanisms by which cells decode frequency-encoded signals remain poorly understood. By reconstructing the cyclic adenosine monophosphate second messenger system in Pseudomonas aeruginosa, we demonstrate that frequency-to-amplitude signal conversion emerges through three distinct filtering modules that decode frequency-encoded signals into gene expression patterns. Our mathematical framework predicts a range of frequency filtering regimes controlled by a dimensionless threshold parameter. We validated these using synthetic circuits and an automated experimental platform. Quantitative analysis reveals that under the given parameter conditions, frequency modulation expands the accessible state space more substantially than amplitude modulation alone. The total number of accessible states scales as the square of the number of regulated genes for frequency-enhanced control, compared with the power of 0.8 for amplitude-only control. This results in approximately two additional bits of information entropy in three-gene systems when using frequency-based control. Our findings establish the fundamental principles of frequency-based signal processing in bacterial second messenger networks, revealing how cells exploit temporal dynamics to regulate multiple genes and expand accessible state spaces. This provides insights into both cellular information physics and design principles for synthetic biology.
DOI: 10.1038/s41567-025-03030-4
Source: https://www.nature.com/articles/s41567-025-03030-4
