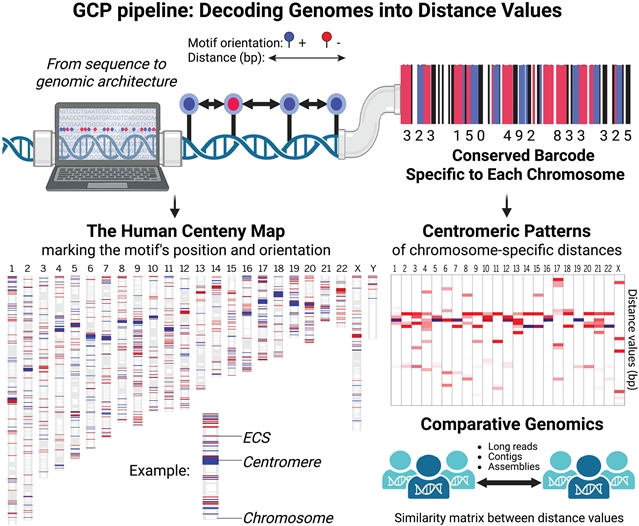
染色体特异性着丝粒模式定义了人类基因组的百年图谱,这一成果由
在这项研究中,该研究团队确定了一个跨越人类基因组的染色体特异性结构模式,由一个功能相关的着丝粒DNA基序的保守间距定义。这些位点沿染色体臂的分布构成了人类的“百岁图谱”。通过主题化cthemtom基因组着丝粒分析(GCP)管道,该研究团队利用基序的位置、取向和组织来构建结构模型,从而实现人类染色体着丝粒的重新分类、着丝粒扩展的检测以及结构变异和错误组装区域的鉴定。从这种模式衍生出的高分辨率图谱不仅为跨越进化和疾病的着丝粒比较分析提供了一个框架,而且为染色体注释、组装和表征提供了一个新的维度。
据介绍,着丝粒在表观遗传上由不同的染色质指定,而它们的DNA在物种和个体之间是不同的。这种广泛的序列差异使着丝粒之间的比较分析具有挑战性。
附:英文原文
Title: Chromosome-specific centromeric patterns define the centeny map of the human genome
Author: Luca Corda, Simona Giunta
Issue&Volume: 2025-07-03
Abstract: Centromeres are epigenetically specified by distinct chromatin, whereas their DNA varies between species and individuals. This extensive sequence divergence makes comparative analyses between centromeres challenging. In this study, we identified a chromosome-specific architectural pattern across the human genome, defined by the conserved spacing of a functionally relevant centromeric DNA motif. The distribution of these sites along chromosome arms constitutes the human “centeny map.” By using a custom Genomic Centromere Profiling (GCP) pipeline, we leveraged the motif’s position, orientation, and organization to construct structural models that enable reclassification of human chromosomal clusters, detection of centromere expansion, and identification of structural variants and misassembled regions. The high-resolution maps derived from this pattern not only provide a framework for comparative analysis of centromeres across evolution and disease but also offer a new dimension for chromosome annotation, assembly, and characterization.
DOI: ads3484
Source: https://www.science.org/doi/10.1126/science.ads3484
