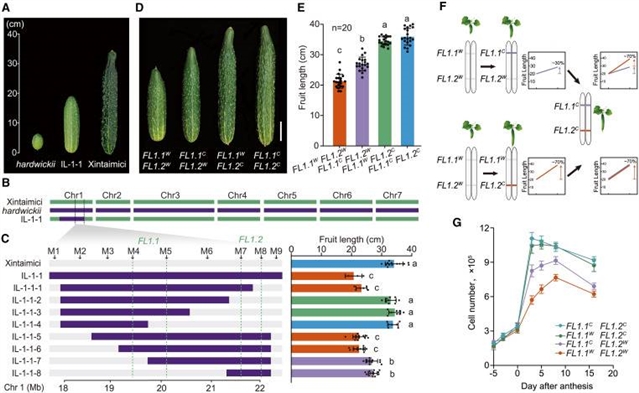
中国农业科学院杨学勇团队报道了同义突变的隐性上位性通过表转录组调控使黄瓜驯化。这一研究成果于2025年7月1日发表在国际顶尖学术期刊《细胞》上。
课题组研究人员发现了两个密切相关的、上位相互作用的基因:YTH1 (RNA N6-甲基腺苷(m6A)解读器)和ACS2(氨基环丙烷-1-羧酸(ACC)合成酶),它们有助于黄瓜果实长度驯化。ACS2的阳性突变是在1287C>T位点的同义词替换。在野生黄瓜中,ACS21287C导致附近腺苷残基的m6A修饰,形成松散的RNA结构构象。YTH1识别m6A修饰,使折叠平衡向最弱的RNA结构构象改变,增加ACS2蛋白水平,导致果实变短。在栽培黄瓜中,ACS21287T破坏m6A甲基化,形成紧凑的RNA结构构象,导致蛋白质产量降低和果实伸长。本研究提供了同义变异通过表转录组调控形成生物学性状的遗传证据。
研究人员表示,同义突变,曾经被称为“沉默”突变,越来越吸引生物学家的兴趣。尽管它们可能影响转录或转录后过程,但它们对生物学性状的影响仍未得到充分研究,特别是在生物体水平上。
附:英文原文
Title: Recessive epistasis of a synonymous mutation confers cucumber domestication through epitranscriptomic regulation
Author: Tongxu Xin, Zhen Zhang, Yueying Zhang, Xutong Li, Shenhao Wang, Guanqun Wang, Haoxuan Li, Bowen Wang, Mengzhuo Zhang, Wenjing Li, Haojie Tian, Zhonghua Zhang, Yu-Lan Xiao, Weixin Tang, Chuan He, Yiliang Ding, Sanwen Huang, Xueyong Yang
Issue&Volume: 2025-07-01
Abstract: Synonymous mutations, once known as “silent” mutations, are increasingly attracting the interest of biologists. Although they may affect transcriptional or post-transcriptional processes, their impact on biological traits remains under-investigated, particularly at the organismal level. Here, we identified two closely linked, epistatically interacting genes: YTH1, an RNA N6-methyladenosine (m6A) reader, and ACS2, an aminocyclopropane-1-carboxylic acid (ACC) synthase, which contribute to cucumber fruit length domestication. The causative mutation in ACS2 is a synonymous substitution at 1287C>T. In wild cucumber, ACS21287C results in m6A modification on nearby adenosine residues and the formation of loose RNA structural conformations. YTH1 recognizes the m6A modification, alters the folding equilibrium toward the weakest RNA structural conformation, and increases the ACS2 protein level, resulting in shorter fruit. In cultivated cucumber, ACS21287T disrupts m6A methylation and forms compact RNA structural conformations, leading to attenuated protein production and fruit elongation. This study provides genetic evidence of synonymous variation shaping a biological trait through epitranscriptomic regulations.
DOI: 10.1016/j.cell.2025.06.007
Source: https://www.cell.com/cell/abstract/S0092-8674(25)00674-9
