美国华盛顿大学Uri Keich团队揭示了利用诱捕法对串联质谱分析中的错误发现率控制进行评估。这一研究成果于2025年6月16日发表在国际顶尖学术期刊《自然—方法学》上。
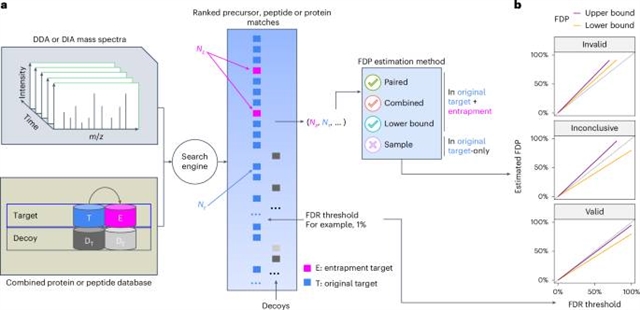
在这里,课题组研究人员确定了验证错误发现率(FDR)控制的三种流行方法:一种无效,一种仅提供下界,一种有效但功率不足。结果是,蛋白质组学社区对实际的FDR控制效果的了解有限,特别是对数据独立获取(DIA)分析。研究小组提出了一个陷阱实验的理论框架,使他们能够严格地表征不同的方法。
此外,该研究组引入了一种更强大的评估方法,并将其与现有技术一起应用于评估现有工具。研究小组首先在更好理解的数据依赖采集设置中验证他们的分析,然后,研究小组分析DIA数据,研究小组发现没有DIA搜索工具始终控制FDR,在单细胞数据集上表现特别差。
据了解,质谱蛋白质组学的一个关键挑战是准确评估错误控制,特别是考虑到软件工具采用不同的方法来报告错误。许多工具都是封闭的,文档记录很差,导致验证策略不一致。
附:英文原文
Title: Assessment of false discovery rate control in tandem mass spectrometry analysis using entrapment
Author: Wen, Bo, Freestone, Jack, Riffle, Michael, MacCoss, Michael J., Noble, William S., Keich, Uri
Issue&Volume: 2025-06-16
Abstract: A critical challenge in mass spectrometry proteomics is accurately assessing error control, especially given that software tools employ distinct methods for reporting errors. Many tools are closed-source and poorly documented, leading to inconsistent validation strategies. Here we identify three prevalent methods for validating false discovery rate (FDR) control: one invalid, one providing only a lower bound, and one valid but under-powered. The result is that the proteomics community has limited insight into actual FDR control effectiveness, especially for data-independent acquisition (DIA) analyses. We propose a theoretical framework for entrapment experiments, allowing us to rigorously characterize different approaches. Moreover, we introduce a more powerful evaluation method and apply it alongside existing techniques to assess existing tools. We first validate our analysis in the better-understood data-dependent acquisition setup, and then, we analyze DIA data, where we find that no DIA search tool consistently controls the FDR, with particularly poor performance on single-cell datasets.
DOI: 10.1038/s41592-025-02719-x
Source: https://www.nature.com/articles/s41592-025-02719-x
Nature Methods:《自然—方法学》,创刊于2004年。隶属于施普林格·自然出版集团,最新IF:47.99
官方网址:https://www.nature.com/nmeth/
投稿链接:https://mts-nmeth.nature.com/cgi-bin/main.plex
