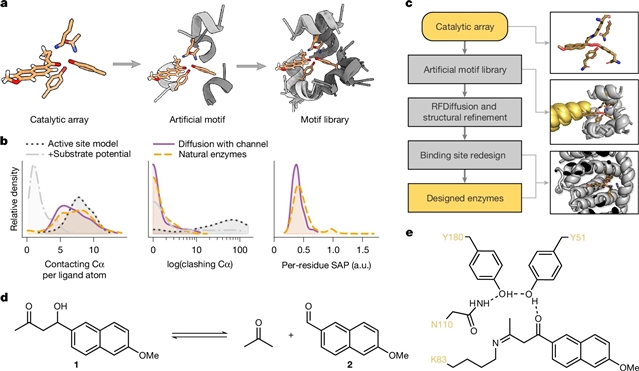
格拉茨工业大学Gustav Oberdorfer团队取得一项新突破。他们开发出基于催化基序支架的计算酶设计。相关论文于2025年12月3日发表在《自然》杂志上。
在这里,该研究团队提出了旋转体倒置片段查找器-扩散(Riff-Diff),这是一种混合机器学习和原子建模策略,用于新蛋白质的脚手架催化阵列。课题组通过设计两种机械上不同的化学转化酶,即反醛醇反应和森田-贝利斯-希尔曼反应,突出了Riff-Diff的一般适用性。研究人员表明,在这两种情况下,有可能产生具有与体外进化优化的催化剂相媲美的活性的催化剂,以及精致的立体选择性。其中六个设计的高分辨率结构揭示了接近原子活性位点的设计精度。原则上,设计策略可以适用于任何催化活性氨基酸阵列。这些发现为从头合成蛋白质催化剂的实际应用奠定了基础,并描述了蛋白质设计和酶催化的基本原理。
据介绍,酶以其优良的选择性、效率和温和的反应条件,在工业和医学上被广泛用作生物催化剂。他们设计的酶可以产生量身定制的生物催化剂,其潜在应用范围超出了自然反应。然而,目前的设计方法需要测试大量的设计,并且大多产生具有低催化活性的新酶。因此,它们需要昂贵的实验优化和高通量筛选才能在体内可行。
附:英文原文
Title: Computational enzyme design by catalytic motif scaffolding
Author: Braun, Markus, Tripp, Adrian, Chakatok, Morakot, Kaltenbrunner, Sigrid, Fischer, Celina, Stoll, David, Bijelic, Aleksandar, Elaily, Wael, Totaro, Massimo G., Moser, Melanie, Hoch, Shlomo Y., Lechner, Horst, Rossi, Federico, Aleotti, Matteo, Hall, Mlanie, Oberdorfer, Gustav
Issue&Volume: 2025-12-03
Abstract: Enzymes find broad use as biocatalysts in industry and medicine owing to their exquisite selectivity, efficiency and mild reaction conditions. Custom-designed enzymes can produce tailor-made biocatalysts with potential applications that extend beyond natural reactions. However, current design methods require testing a large number of designs and mostly produce de novo enzymes with low catalytic activities1,2,3. As a result, they require costly experimental optimization and high-throughput screening to be industrially viable4,5. Here we present rotamer inverted fragment finder–diffusion (Riff-Diff), a hybrid machine learning and atomistic modelling strategy for scaffolding catalytic arrays in de novo proteins. We highlight the general applicability of Riff-Diff by designing enzymes for two mechanistically distinct chemical transformations, the retro-aldol reaction and the Morita–Baylis–Hillman reaction. We show that in both cases, it is possible to generate catalysts that exhibit activities rivalling those optimized by in vitro evolution, along with exquisite stereoselectivity. High-resolution structures of six of the designs revealed near-atomic active site design precision. The design strategy can, in principle, be applied to any catalytically competent amino acid array. These findings lay the basis for practical applicability of de novo protein catalysts in synthesis and describe fundamental principles of protein design and enzyme catalysis.
DOI: 10.1038/s41586-025-09747-9
Source: https://www.nature.com/articles/s41586-025-09747-9
官方网址:http://www.nature.com/
