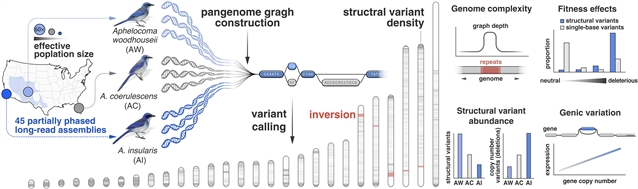
利用45个长读从头基因组组合和泛基因组工具,该课题组分析了3个近缘种的北美松鸦(黑斑松鸦和灌木松鸦)的SVs,它们的有效种群大小范围为55倍。该课题组研究人员发现基因组结构的快速进化,包括在复杂卫星景观变化的驱动下,基因组大小减少了约100兆碱基。受变异长度和种群大小的调节,SVs表现出轻微的衰减动态,只有在最大的种群中才有一致的适应性固定证据。基因拷贝数变异与群体大小呈反比关系,表明它们具有强烈的破坏动力学,并对基因表达产生影响。他们的长读数据集和泛基因组分析证明了种群大小如何影响基因组的复杂性。
据悉,结构变异(SVs)在脊椎动物基因组中广泛存在,但其进化动力学仍然知之甚少。
附:英文原文
Title: Multispecies pangenomes reveal a pervasive influence of population size on structural variation
Author: Scott V. Edwards, Bohao Fang, Danielle Khost, George E. Kolyfetis, Rebecca G. Cheek, Devon A. DeRaad, Nancy Chen, John W. Fitzpatrick, John E. McCormack, W. Chris Funk, Cameron K. Ghalambor, Erik Garrison, Andrea Guarracino, Heng Li, Timothy B. Sackton
Issue&Volume: 2025-12-11
Abstract: Structural variants (SVs) are widespread in vertebrate genomes, yet their evolutionary dynamics remain poorly understood. Using 45 long-read de novo genome assemblies and pangenome tools, we analyze SVs among three closely related species of North American jays (Aphelocoma, scrub-jays) displaying a 55-fold range in effective population size. We find rapid evolution of genome architecture, including ~100-megabase decreases in genome size driven by shifts in complex satellite landscapes. SVs exhibit slightly deleterious dynamics modulated by variant length and population size, with consistent evidence of adaptive fixation only in the largest population. Gene copy number variants exhibit an inverse relationship with population size, indicating strongly deleterious dynamics, with consequences for gene expression. Our long-read dataset and pangenome analysis demonstrate how population size shapes genome complexity.
DOI: adw1931
Source: https://www.science.org/doi/10.1126/science.adw1931
