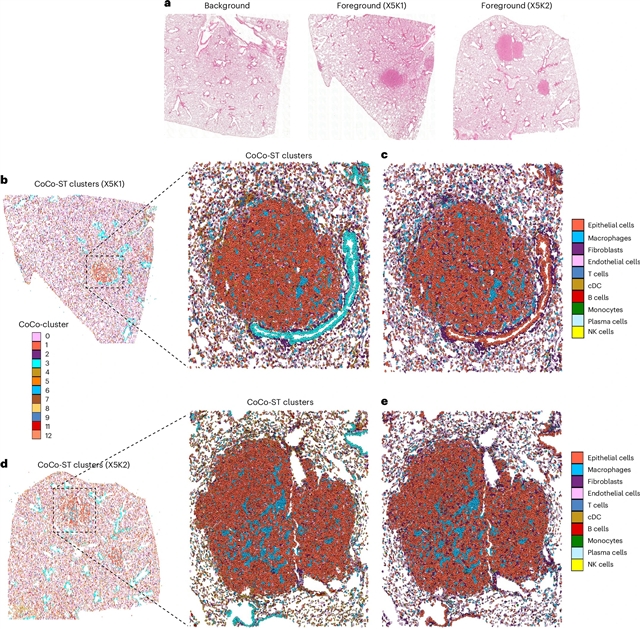
德克萨斯大学吴佳团队宣布他们的研究显示,CoCo-ST在空间转录组学数据集中检测全局和局部生物结构。2025年10月13日出版的《自然—细胞生物学》发表了这项成果。
为了解决这个问题,该课题组人员引入了“比较和对比空间转录组学”(CoCo-ST),这是一种图形对比特征表示框架。通过将目标样本与背景样本进行比较,CoCo-ST可以检测到高方差、广泛共享的结构和低方差、组织特异性的特征。它具有技术优势,包括多样本集成、批处理效果校正以及从现场级Visium数据到单细胞Xenium Prime 5K和亚细胞Visium HD数据的可扩展性。
研究小组将CoCo-ST与十种最先进的空间域检测算法进行了基准测试,这些算法以无主题肺癌前病变样本为主题,证明其识别低方差空间结构的卓越能力被其他方法所忽视。CoCo-ST还能有效区分Visium HD和Xenium Prime 5K数据中的细胞密度和生态位结构。CoCo-ST可在GitHub (https://github.com/WuLabMDA/CoCo-ST)访问。
据了解,空间域检测方法通常关注高变异结构,如肿瘤邻近区域基因表达变化剧烈,而忽略低变异结构,基因表达变化微妙,如邻近正常和早期腺瘤区域之间的结构。
附:英文原文
Title: CoCo-ST detects global and local biological structures in spatial transcriptomics datasets
Author: Aminu, Muhammad, Zhu, Bo, Vokes, Natalie, Chen, Hong, Hong, Lingzhi, Li, Jianrong, Fujimoto, Junya, Chaib, Mehdi, Yang, Yuqiu, Wang, Bo, Poteete, Alissa, Nilsson, Monique B., Le, Xiuning, Cascone, Tina, Jaffray, David, Navin, Nicholas, Wang, Tao, Byers, Lauren A., Gibbons, Don L., Heymach, John, Chen, Ken, Cheng, Chao, Zhang, Jianjun, Wu, Jia
Issue&Volume: 2025-10-13
Abstract: Spatial domain detection methods often focus on high-variance structures, such as tumour-adjacent regions with sharp gene expression changes, while missing low-variance structures with subtle gene expression shifts, like those between adjacent normal and early adenoma regions. Here, to address this, we introduce ‘compare and contrast spatial transcriptomics’ (CoCo-ST), a graph contrastive feature representation framework. By comparing a target sample with a background sample, CoCo-ST detects both high-variance, broadly shared structures and low-variance, tissue-specific features. It offers technical advantages, including multisample integration, batch-effect correction and scalability across technologies from spot-level Visium data to single-cell Xenium Prime 5K and subcellular Visium HD data. We benchmarked CoCo-ST against ten state-of-the-art spatial-domain-detection algorithms using mouse lung precancerous samples, demonstrating its superior ability to identify low-variance spatial structures overlooked by other methods. CoCo-ST also effectively distinguishes cell clusters and niche structures in Visium HD and Xenium Prime 5K data. CoCo-ST is accessible at GitHub (https://github.com/WuLabMDA/CoCo-ST).
DOI: 10.1038/s41556-025-01781-z
Source: https://www.nature.com/articles/s41556-025-01781-z
Nature Cell Biology:《自然—细胞生物学》,创刊于1999年。隶属于施普林格·自然出版集团,最新IF:28.213
官方网址:https://www.nature.com/ncb/
投稿链接:https://mts-ncb.nature.com/cgi-bin/main.plex
