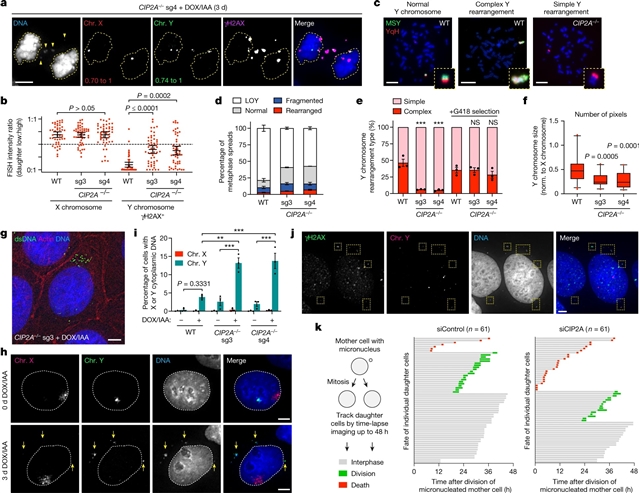
美国德克萨斯大学Peter Ly等研究人员合作揭示来自微核的粉碎性染色体的有丝分裂聚集。这一研究成果于2023年5月10日在线发表在国际学术期刊《自然》上。
研究人员用活细胞成像技术跟踪了微核染色体,并显示在整个有丝分裂过程中,无着丝粒片段在空间上紧密相邻,由单个子细胞不对称地继承。从机制上讲,CIP2A-TOPBP1复合物在间期与破裂的微核内的DNA损伤过早结合,这使碎裂的染色体在进入有丝分裂时准备好聚在一起。CIP2A-TOPBP1的失活导致无着丝粒片段分散在整个有丝分裂的细胞质中,随机地分配到两个子细胞的细胞核中,并异常地作为细胞质DNA错误累积。
有丝分裂的聚集有利于将无着丝粒片段重新组装成重新排列的染色体,而这些染色体缺乏典型的染色体衰退所特有的大量DNA拷贝数损失。对泛癌基因组的综合分析显示,在不同的肿瘤类型中,DNA拷贝数中性重排的聚集,称为平衡染色体碎裂,导致了已知癌症驱动事件的获得。因此,不同的染色体剥离模式可以用微核中碎裂染色体的空间聚集来解释。
研究人员表示,复杂的基因组重排可由困在微核内的错位染色体的灾难性粉碎产生,这一过程被称为染色体碎裂。由于每条染色体都含有一个中心粒,目前仍不清楚在有丝分裂过程中,破碎的染色体所产生的无着丝粒片段是如何在子细胞之间继承的。
附:英文原文
Title: Mitotic clustering of pulverized chromosomes from micronuclei
Author: Lin, Yu-Fen, Hu, Qing, Mazzagatti, Alice, Valle-Incln, Jose Espejo, Maurais, Elizabeth G., Dahiya, Rashmi, Guyer, Alison, Sanders, Jacob T., Engel, Justin L., Nguyen, Giaochau, Bronder, Daniel, Bakhoum, Samuel F., Corts-Ciriano, Isidro, Ly, Peter
Issue&Volume: 2023-05-10
Abstract: Complex genome rearrangements can be generated by the catastrophic pulverization of missegregated chromosomes trapped within micronuclei through a process known as chromothripsis1,2,3,4,5. As each chromosome contains a single centromere, it remains unclear how acentric fragments derived from shattered chromosomes are inherited between daughter cells during mitosis6. Here we tracked micronucleated chromosomes with live-cell imaging and show that acentric fragments cluster in close spatial proximity throughout mitosis for asymmetric inheritance by a single daughter cell. Mechanistically, the CIP2A–TOPBP1 complex prematurely associates with DNA lesions within ruptured micronuclei during interphase, which poises pulverized chromosomes for clustering upon mitotic entry. Inactivation of CIP2A–TOPBP1 caused acentric fragments to disperse throughout the mitotic cytoplasm, stochastically partition into the nucleus of both daughter cells and aberrantly misaccumulate as cytoplasmic DNA. Mitotic clustering facilitates the reassembly of acentric fragments into rearranged chromosomes lacking the extensive DNA copy-number losses that are characteristic of canonical chromothripsis. Comprehensive analysis of pan-cancer genomes revealed clusters of DNA copy-number-neutral rearrangements—termed balanced chromothripsis—across diverse tumour types resulting in the acquisition of known cancer driver events. Thus, distinct patterns of chromothripsis can be explained by the spatial clustering of pulverized chromosomes from micronuclei.
DOI: 10.1038/s41586-023-05974-0
Source: https://www.nature.com/articles/s41586-023-05974-0
Nature:《自然》,创刊于1869年。隶属于施普林格·自然出版集团,最新IF:69.504
官方网址:http://www.nature.com/
投稿链接:http://www.nature.com/authors/submit_manuscript.html
