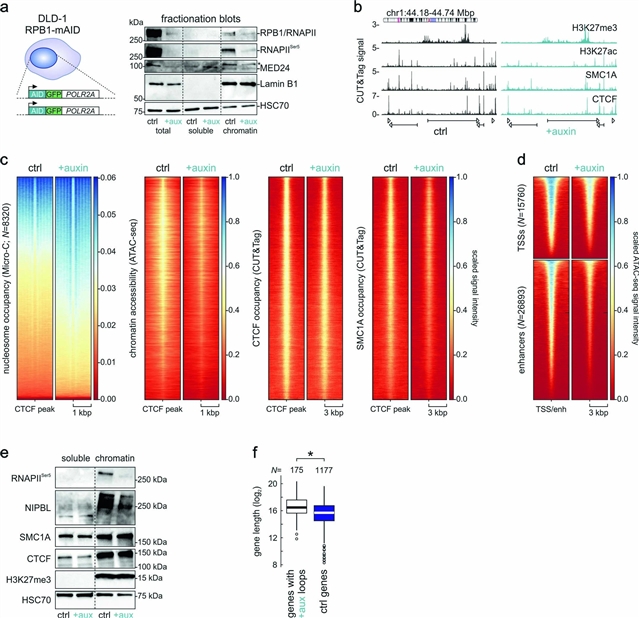
德国哥廷根大学Argyris Papantonis团队发现,增强子-启动子接触的形成需要RNAPII并拮抗环的挤压。相关论文于2023年4月3日在线发表于国际学术期刊《自然—遗传学》。
研究人员测试了RNA聚合酶II(RNAPII)在一个细胞系统中不同规模的间期染色质组织中的作用,这个系统允许其快速、由生长素介导的降解。研究人员结合Micro-C和计算模型来描述了RNAPII耗尽后获得或失去的不同环路的子集。获得的环路,其挤压被RNAPII拮抗,几乎无一例外地通过参与新的或重新连接的CTCF锚而形成。丢失的环路选择性地影响了由RNAPII锚定的增强子和启动子之间的接触,并解释了对大多数基因的压制。
令人惊讶的是,启动子与启动子之间的相互作用基本上不受聚合酶耗竭的影响,而且聚合酶的占用是持续的。总之,这些发现使RNAPII在转录中的作用与它直接参与建立基因组范围内的调节性三维染色质接触相协调,同时也揭示了对黏着素环挤压的影响。
据了解,同型染色质相互作用和环状挤压被认为是哺乳动物染色体折叠的两个主要驱动因素。
附:英文原文
Title: Enhancer–promoter contact formation requires RNAPII and antagonizes loop extrusion
Author: Zhang, Shu, belmesser, Nadine, Barbieri, Mariano, Papantonis, Argyris
Issue&Volume: 2023-04-03
Abstract: Homotypic chromatin interactions and loop extrusion are thought to be the two main drivers of mammalian chromosome folding. Here we tested the role of RNA polymerase II (RNAPII) across different scales of interphase chromatin organization in a cellular system allowing for its rapid, auxin-mediated degradation. We combined Micro-C and computational modeling to characterize subsets of loops differentially gained or lost upon RNAPII depletion. Gained loops, extrusion of which was antagonized by RNAPII, almost invariably formed by engaging new or rewired CTCF anchors. Lost loops selectively affected contacts between enhancers and promoters anchored by RNAPII, explaining the repression of most genes. Surprisingly, promoter–promoter interactions remained essentially unaffected by polymerase depletion, and cohesin occupancy was sustained. Together, our findings reconcile the role of RNAPII in transcription with its direct involvement in setting-up regulatory three-dimensional chromatin contacts genome wide, while also revealing an impact on cohesin loop extrusion.
DOI: 10.1038/s41588-023-01364-4
Source: https://www.nature.com/articles/s41588-023-01364-4
Nature Genetics:《自然—遗传学》,创刊于1992年。隶属于施普林格·自然出版集团,最新IF:41.307
官方网址:https://www.nature.com/ng/
投稿链接:https://mts-ng.nature.com/cgi-bin/main.plex
