近日,中国科学院分子植物科学卓越创新中心张余等研究人员合作揭示固有转录终止的结构基础。2023年1月11日,《自然》杂志在线发表了这项成果。
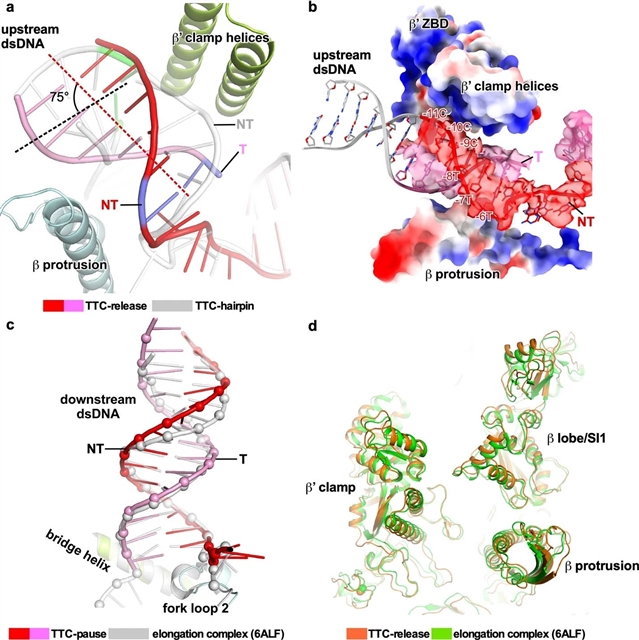
研究人员报告了一组大肠杆菌转录固有终止复合物的单粒子冷冻电镜结构,这代表了该事件的关键中间状态。这些结构显示了RNA聚合酶如何在终止子序列处暂停,终止子RNA发夹如何在RNA聚合酶内折叠,以及RNA聚合酶如何倒转转录泡来释放RNA,然后释放DNA。这些大分子快照确定了细菌固有终止的结构机制,以及与所有RNA聚合酶的因子非依赖终止有关的RNA释放和DNA塌陷的途径。
据悉,在所有生物体中,基因转录都需要高效和准确的终止。细菌和真核生物的细胞RNA聚合酶都可以通过一种不依赖因子的终止途径来终止其转录,在细菌中称为固有终止转录,其中RNA聚合酶识别终止子序列,停止核苷酸的添加,并自发释放新生RNA。
附:英文原文
Title: Structural basis for intrinsic transcription termination
Author: You, Linlin, Omollo, Expery O., Yu, Chengzhi, Mooney, Rachel A., Shi, Jing, Shen, Liqiang, Wu, Xiaoxian, Wen, Aijia, He, Dingwei, Zeng, Yuan, Feng, Yu, Landick, Robert, Zhang, Yu
Issue&Volume: 2023-01-11
Abstract: Efficient and accurate termination is required for gene transcription in all living organisms1,2. Cellular RNA polymerases in both bacteria and eukaryotes can terminate their transcription through a factor-independent termination pathway3,4—called intrinsic termination transcription in bacteria—in which RNA polymerase recognizes terminator sequences, stops nucleotide addition and releases nascent RNA spontaneously. Here we report a set of single-particle cryo-electron microscopy structures of Escherichia coli transcription intrinsic termination complexes representing key intermediate states of the event. The structures show how RNA polymerase pauses at terminator sequences, how the terminator RNA hairpin folds inside RNA polymerase, and how RNA polymerase rewinds the transcription bubble to release RNA and then DNA. These macromolecular snapshots define a structural mechanism for bacterial intrinsic termination and a pathway for RNA release and DNA collapse that is relevant for factor-independent termination by all RNA polymerases.
DOI: 10.1038/s41586-022-05604-1
Source: https://www.nature.com/articles/s41586-022-05604-1
Nature:《自然》,创刊于1869年。隶属于施普林格·自然出版集团,最新IF:69.504
官方网址:http://www.nature.com/
投稿链接:http://www.nature.com/authors/submit_manuscript.html
