原文链接:https://marketing.springernature.com/SAP/CUAN/ZCUAN_PERSEMAIL?sap-outbound-id=0000011883:1:26504
微信链接:https://mp.weixin.qq.com/s/Ue3WjawIXL0I8ueqd_kJ1A
Genome Biology很高兴与您分享本刊近期的亮点文章、征稿信息和期刊新闻!
精选文集
点击此处,阅读文集。
Microbiome Biology 特刊正式发行
Genome Medicine 和BMC Biology 共同推出的微生物组生物学的特刊已经正式发行了!本特刊的客座编辑为来自美国加州大学圣地亚哥分校的Rob Knight 教授、宾夕法尼亚大学的Elizabeth Grice 博士、德国蒂宾根大学发育生物学的Ruth Ley博士和来自比利时鲁汶大学的Jeroen Raes 博士。本特刊涵盖微生物学各个领域的研究、方法和评论文章。
亮点文章
点击此处,阅读论文。

Mechanism of a COMPASS-like complex in Arabidopsis
拟南芥中COMPASS样复合物的作用机制
Doi:10.1186/s13059-019-1705-4
Anne-Sophie Fiorucci, Clara Bourbousse et al.
来自瑞士洛桑大学的Fiorucci 等人研究发现:COMPASS样复合物可以利用H2B独立的单泛素通路来介导拟南芥中H3K4me3的沉积。
点击此处,阅读论文。

Biological significance of the 13q12.12 locus in lung cancer risk
13q12.12位点在肺癌患病风险中的生物学意义
Doi:10.1186/s13059-019-1696-1
Lipei Shao, Xianglin Zuo et al.
来自中国南京医科大学的Lipei Shao等人探讨了13q12.12位点是如何与肺癌的患病风险息息相关的,他们的研究揭示了可能存在一种新型的对p53有应答的增强子参与到上述作用机制中。
点击此处,阅读论文。
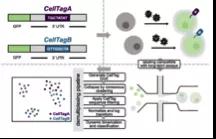
CellTag Indexing: genetic barcode-based sample multiplexing for single-cell genomics
CellTag索引:将基于遗传条形码的样本复用于单细胞基因组中
Doi:10.1186/s13059-019-1699-y
Chuner Guo, Wenjun Kong et al.
来自华盛顿大学圣路易斯医学院的研究者发现了一种新的单细胞研究方法,可以利用预定义的遗传条形码在体外和体内随时间变化对混合样本进行标记和跟踪。
其他内容
Next-generation genome annotation: we still struggle to get it right
Steven L. Salzberg
Doi: 10.1186/s13059-019-1715-2
At the forefront of the sequencing revolution—notes from the RNGS19 conference
Sander Wuyts and Nicola Segata
Doi: 10.1186/s13059-019-1714-3
One reference genome is not enough
Xiaofei Yang et. al.
Doi: 10.1186/s13059-019-1717-0
Mouse microbiomes: overlooked culprits of experimental variability
Maria-Luisa Alegre
Doi: 10.1186/s13059-019-1723-2
Genome-wide analysis of DNA replication timing in single cells: Yes! We’re all individuals
Anne D. Donaldson and Conrad A. Nieduszynski
Doi: 10.1186/s13059-019-1719-y
Genomics and data science: an application within an umbrella
Fábio C. P. Navarro, et al.
Doi: 10.1186/s13059-019-1724-1
Addressing confounding artifacts in reconstruction of gene co-expression networks
Princy Parsana et al.
Doi: 10.1186/s13059-019-1700-9
Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches
Gabriele Andrea Lugli et al.
Doi: 10.1186/s13059-019-1711-6
Human cleaving embryos enable robust homozygotic nucleotide substitutions by base editors
Meiling Zhang et. al.
Doi: 10.1186/s13059-019-1703-6
VULCAN integrates ChIP-seq with patient-derived co-expression networks to identify GRHL2 as a key co-regulator of ERa at enhancers in breast cancer
Andrew N. Holding et al.
Doi: 10.1186/s13059-019-1698-z
Genomic signatures accompanying the dietary shift to phytophagy in polyphagan beetles
Mathieu Seppey et al.
Doi: 10.1186/s13059-019-1704-5
Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight
Mark T. W. Ebbert et al.
Doi: 10.1186/s13059-019-1707-2
Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex
Zelin Liu et al.
Doi: 10.1186/s13059-019-1701-8
SCRABBLE: single-cell RNA-seq imputation constrained by bulk RNA-seq data
Tao Peng et al.
Doi: 10.1186/s13059-019-1681-8
ChiCMaxima: a robust and simple pipeline for detection and visualization of chromatin looping in Capture Hi-C
Yousra Ben Zouari et. al.
Doi: 10.1186/s13059-019-1706-3
WhoGEM: an admixture-based prediction machine accurately predicts quantitative functional traits in plants
Laurent Gentzbittel et al.
Doi: 10.1186/s13059-019-1697-0
OSCA: a tool for omic-data-based complex trait analysis
Futao Zhang et al.
Doi: 10.1186/s13059-019-1718-z
Correction to: Functional variation in allelic methylomes underscores a strong genetic contribution and reveals novel epigenetic alterations in the human epigenome
Warren A Cheung et al.
Doi: 10.1186/s13059-019-1702-7
Comments on the model parameters in “SiFit: inferring tumor trees from single-cell sequencing data under finite-sites models”
Hamim Zafar et al.
Doi: 10.1186/s13059-019-1692-5
阅读英文全文请访问:
https://marketing.springernature.com/SAP/CUAN/ZCUAN_PERSEMAIL?sap-outbound-id=0000011883:1:26504
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。